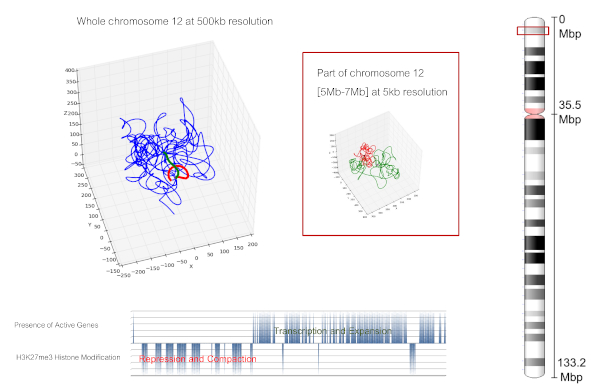
The python-code ChromStruct provides an estimate of the 3D structure of the chromatin fibre in cell nuclei from the contact frequency data produced by a "Chromosome conformation capture" experiment (information on DNA contacts on populations of millions of cells).
The algorithm adopts a multiscale approach to explore the structure of chromosomes at different resolution levels and has a Bayesian optimization function to explore the space of the possible configurations that the chromatin fiber can assume within the nucleus compatibly with the constraints and experimental data. The code is open source and equipped with a GUI through which it is possible to set all the free parameters.
The fiber is initially divided into independent segments, whose conformations are evaluated separately and then modeled as single elements of a lower resolution fiber. The algorithm proceeds iteratively until the minimum possible resolution is reached. ChromStruct requires as input only the matrix of the DNA contact frequencies and returns as output the coordinates of the chain at full resolution, in txt and pdb format.
References:
Caudai C, Salerno E, Zoppe M, Tonazzini A. Estimation of the Spatial Chromatin Structure Based on a Multiresolution Bead-Chain Model. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2019;16(2):550-559.
Caudai C, Salerno E, Zoppe M, Merelli I, Tonazzini A. ChromStruct 4: A Python Code to Estimate the Chromatin Structure from Hi-C Data. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2019; 16: 1867-1878.
Caudai C, Salerno E, Zoppè M, Tonazzini A. Inferring 3D chromatin structure using a multiscale approach based on quaternions. BMC Bioinformatics. 2015;16:234.
Software:
E. Salerno, C. Caudai. CHROMSTRUCT v4.2 - Reconstruction of 3D chromatin structure from chromosome conformation capture data. Software, 2018, CNR-ISTI, Pisa, 2018-388694 DOI: 10.13140/RG.2.2.26123.39208.
Immagini: